| Home | Research | News | Papers | Software | People | Links | Contact |
| RESEARCH
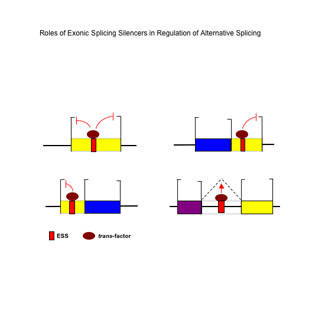
Christopher B. Burge Whitehead Career Development Associate Professor Room 68-223 Burge Lab Website: http://genes.mit.edu/burgelab OVERVIEW Computational Biology of Gene Expression. We study mechanisms of posttranscriptional gene regulation using a combination of computational and experimental methods. A long-term goal is to understand the RNA splicing code: how the precise locations of exons and splice sites are identified in primary transcripts, and how this code is altered in cell- and condition-specific alternative splicing. Current efforts are focused on identifying splicing cis-regulatory elements and associated splicing factors, and understanding the context-dependent activities and functional interactions between these elements. We also study the roles that microRNAs (miRNAs) play in gene regulation, with an emphasis on determining the rules for miRNA-directed targeting of mRNAs. We are beginning to study the relationship between alternative cleavage and polyadenylation, which is commonly used to generate alternative mRNA isoforms differeing in their 3' UTRs, and miRNA regulation. RESEARCH SUMMARY Splicing Regulatory Elements and Interactions. The RNA splicing machinery is directed to particular locations in RNA transcripts by exon and intron sequences that either enhance or silence splicing at nearby splice sites. We have developed computational approaches to identify splicing enhancer elements that are active in mammalian cells, and found that these elements are predictive of splicing phenotypes of mutations in human genes. We are exploring the evolution of splicing enhancers to understand the extent to which different classes of enhancers have distinct or interchangeable functions. To discover silencers of splicing, we have developed a cell fluorescence-based screening method, and used this approach to identify a diverse collection of exonic splicing silencer (ESS) motifs. Almost all of these elements share a consistent pattern of context-dependent activity, silencing splicing when present in exons, activating splicing when present in introns, and altering splice site choice when present between competing splice sites. We are exploring the roles that ESSs play in a variety of types of regulated alternative splicing events, using comparative genomic and molecular approaches, and integrating ESS and other motifs into algorithms that simulate exon recognition by the splicing machinery. Reverse genetics (RNAi) is being used to identify splicing factors required for the activity of these ESS motifs. |
|||
|---|---|---|---|
Figure 6. A model for the activities of exonic splicing silencers in splicing regulation. (From Wang et al. Mol. Cell 23, 61-70, 2006.) |
|||
Core splice site motifs and splicing regulatory elements function together to regulate splicing decisions. We are beginning to explore the functional interactions between classes of splicing regulatory elements, using genomic/comparative genomic analyses together with splicing reporter assays. One direction involves identifying pairs of elements that preferentially occur together in adjacent gene regions. This approach has led to the identification of a pair of intronic motifs that can function cooperatively to silence the splicing of intervening exons. Pairs of co-occurring motifs in other gene regions are also being explored. Patterns of evolutionary compensation between different classes of splicing regulatory elements can also provide clues to functional interactions. We are exploring in depth a pattern in which intronic splicing enhancers (ISEs) enhance the splicing of exons whose 5' splice sites have moderate intrinsic strength much more strongly than for other exons whose splice sites are either weaker or stronger. This splice site strength-specific activity suggests unexpected complexity in the activity of splicing enhancers. To explore the regulatory functions of alternative splicing in mammalian differentiation, in collaboration with the Phillip Sharp lab we are using splicing-sensitive microarrays to analyze changes in the expression of alternative mRNA isoforms following activation of primary T-cells. MicroRNA targeting. In animals, mRNAs are frequently targeted for repression by miRNAs. In collaboration with the David Bartel lab, we have used comparative genomics approaches to explore miRNA targeting, leading to development of the TargetScan and TargetScanS miRNA target prediction algorithms. These studies pointed to the critical importance for miRNA targeting of ‘seed match’ segments complementary to miRNA nucleotides 2-7, dubbed the miRNA ‘seed’ region. They also showed that at least one-third of human genes represent conserved targets of miRNAs. The predicted targets of some miRNA families have clear functional relatedness, and certain miRNAs and miRNA clusters appear to preferentially target tumor suppressor and other genes involved in growth control, supporting potential roles for miRNAs in cancer. In addition to promoting translational inhibition of mRNAs, miRNAs often direct accelerated decay of targeted mRNAs, so that changes in mRNA as well as protein levels can be used as a readout of targeting. We are exploring targeting rules using available mRNA array data from miRNA/siRNA transfection experiments, as well as our own data from mouse cells following knockout of the miRNA processing enzyme, Dicer. These studies have led to the idea that there are different types of seed matches that confer different degrees of miRNA-directed repression. These studies have also found evidence that certain positions in the targeted mRNA may be recognized directly by protein compo-nents of the silencing complex rather than through pairing to the miRNA. The Dicer knockout data allow us to assess the degree to which mRNA targeting by endogenous miRNAs follows similar rules to targeting by exogenously added miRNAs or siRNAs. Current efforts are focused on identifying additional mRNA features that enhance or inhibit miRNA-directed targeting. SELECTED PUBLICATIONSWang, Z., Xiao, X., Van Nostrand, E. and Burge, C. B. (2006). General and specific functions of exonic splicing silencers in splicing control. Mol. Cell 23, 61-70. Yeo, G., Van Nostrand, E., Holste, D., Poggio, T. and Burge, C. B. Identification and analysis of alternative splicing events conserved between human and mouse. Proc. Natl. Acad. Sci USA 102, 2850-2855 (2005). Lewis, B., Burge, C. B., and Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates thousands of human microRNA targets. Cell 120, 15-20 (2005). Wang, Z., Rolish, M., Yeo, G., Tung, V., Mawson, M. and Burge, C. B. Systematic identification and analysis of exonic splicing silencers. Cell 119, 831-845 (2004). Nielsen, C., Friedman, B., Birren, B., Burge, C. B. and Galagan, J. Patterns of intron gain and loss in fungi. PLoS Biol. 2, e422 (2004). Fairbrother, W. G., Holste, D., Burge, C. B. and Sharp, P. A. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2, e268 (2004). Lewis, B. P., Shih, I-h., Jones-Rhoades, M. W., Bartel, D. P. and Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787-798 (2003). Lim, L. P., Glasner, M., Yekta, S., Burge, C. B. and Bartel, D. P. Vertebrate microRNA genes. Fairbrother, W., Yeh, R.-F.,Sharp, P. A. and Burge, C. B. Predictive identification of exonic
|
|||
|
|||